Modern Genomics II
I cannot understand his accent...
Contents that (I think) are important
Linkage disequilibrium (LD)
Coefficient of linkage disequilibrium \( D \)
Given two loci \(A\) and \(B\) (can have allele \(A\) or \(a\), \(B\) or \(b\)), the coefficient of linkage disequilibrium \(D\) is defined as
\[ D = \frac{P_{AB} - P_A P_B}{P_A P_B} \]
Below shows two examples of \(D\) calculation.
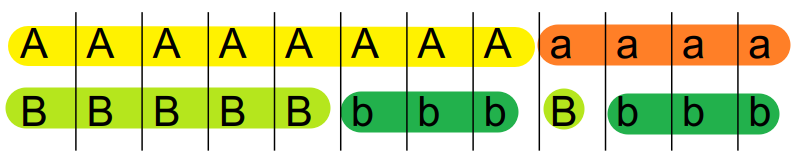
\[ D = \frac{P_{AB} - P_{A} P_{B}}{P_{A} P_{B}} = \frac{5}{12} - \frac{8}{12}\times\frac{6}{12} = \frac{1}{12} \]
A and B are linked to some extent.
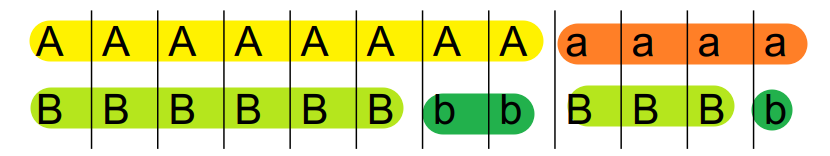
\[ D = \frac{P_{AB} - P_{A} P_{B}}{P_{A} P_{B}} = \frac{6}{12} - \frac{8}{12}\times\frac{9}{12} = 0 \]
Here A and B are not linked at all.
Basically we are testing whether P(A) and P(B) are independent.
Relative measures of LD
- \(r^2 = \frac{D}{P_A P_a P_B P_b}\)
- \(D' = \frac{D}{D_{\max}}\) if \(D\) is positive, \(D' = \frac{D}{D_{\min}}\) if \(D\) is negative
Nucleotide diversity \(\pi\)
Average proportion of pairwise differences between the sequences
Give a multiple sequence alignment, \(\pi\) is defined as
\[ \pi = \sum_{i < j} \frac{\pi_{ij}}{n_c} \]
where \(n_c = \frac{n(n-1)}{2}\) is the number of pairwise comparisons, and \(\pi_{ij}\) is the proportion of differences between the \(i\)th and \(j\)th sequences.
Below shows an example of \(\pi\) calculation.

\[ \pi = \sum_{i < j} \frac{\pi_{ij}}{n_c} = \frac{\pi_{12} + \pi_{13} + \pi_{23}}{3} = 0.2 \]
Basically just calculating the average number of differences in the matrix.
Course contents according to recording
Title: Applications of next-generation sequencing and other genomic techniques
- Resequencing to identify genetic basis of phenotypic variation
- Outlier approach
- Genome-wide association studies (GWAS) and linkage disequilibrium
- Introduction to RNA-seq (transcriptome) focusing on genome duplication
- Resequencing to detect signature of selection: nucleotide diversity
Microbial community and metagenomics, ChIP-seq, epigenomics, whole genome assembly as reference
- Evolutionary functional genomics can...
- identify ecologically relevant genes
- inferring selection & population processes